New more efficient method of sampling biodiversity showcased in major UK estuaries
 Some of the diverse species, invisible to the human eye, found in the Thames and Mersey estuaries seen down the microscope.Two of the UK’s major estuaries have proved to be a successful testing-ground for an effective new method of ‘health-checking’ aquatic biodiversity, which could lead to faster and more efficient sampling for other sites.
Some of the diverse species, invisible to the human eye, found in the Thames and Mersey estuaries seen down the microscope.Two of the UK’s major estuaries have proved to be a successful testing-ground for an effective new method of ‘health-checking’ aquatic biodiversity, which could lead to faster and more efficient sampling for other sites.
“Bio-monitoring” or assessing the impacts of human activities in the natural environment is often achieved by monitoring biological diversity. Existing methods rely on manual identification, but that takes time, resources and often focuses on larger creatures, that sometimes may not be able to reflect accurately the health of particular habitats.
Investigators from Bangor University have shown that by focusing on DNA obtained from smaller animals, you can detect distinct ecological patterns of biodiversity from key UK estuaries. The work on the Thames and Mersey estuaries performed in collaboration with the UK Environment Agency, showcases the efficiency of using genetic sequence data for bio-monitoring, but also the effect of local factors (e.g. geography or pollution history) in determining biodiversity patterns.
“Because we know so little about the effects of human activities and man-made chemicals in the natural environment, measuring ecosystem health is often performed by measuring the diversity of particular biological indicator species,” explains Simon Creer of the School of Biological Sciences, corresponding author of a recent paper in ISME, the Multidisciplinary Journal of Microbial Ecology.
“Bio-indicator species reflect particular environmental conditions, but only represent the tip of the iceberg of the 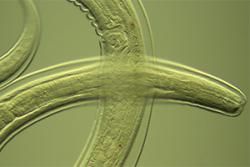 potential biodiversity that could be used for understanding environmental health. By looking at a broader range of species, we may be able to understand better how man-made chemicals are influencing our natural environment and improve ecosystem health.”
potential biodiversity that could be used for understanding environmental health. By looking at a broader range of species, we may be able to understand better how man-made chemicals are influencing our natural environment and improve ecosystem health.”
In the study, a team led by investigators from the University's College of Natural Sciences wanted to find out what natural factors affected the diversity of microscopic creatures, by focusing on environmental DNA (eDNA, obtained from thousands of individual organisms) of estuarine sediment cores.
Following sampling and processing of the sediments the first author of the study, Dr. Delphine Lallias, was able to visualize the biodiversity of thousands of species of animals, plants, fungi and other microscopic organisms along the full salinity range of the Thames and Mersey estuaries. By doing so, the team were also able to prove for the first time, through this quantitative analysis, that the diversity of different groups of organisms were affected by different environmental factors in the two estuaries.
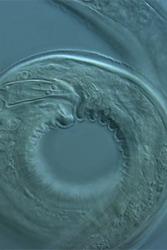 The combined discoveries show the potential for using new technologies for more effective bio-monitoring and suggest that either local, geographic or anthropogenic effects were likely responsible for the resulting compositions of the different biological communities.
The combined discoveries show the potential for using new technologies for more effective bio-monitoring and suggest that either local, geographic or anthropogenic effects were likely responsible for the resulting compositions of the different biological communities.
Dr. Simon Creer commented:
“One of the advances provided by the project is the sheer volume of work that can be undertaken by a modest team, equipped with a molecular biology laboratory. In the study, in a period of months we catalogued the biodiversity of over 20 different groups of microscopic organisms from over 100 samples, many of which are likely to be new to science. To repeat the work using morphological identification would take a large team of researchers many years.”
The team, along with other groups in the UK are working on a variety of projects with the Environment Agency aiming to enhance existing monitoring approaches.
The work, funded by the Natural Environment Research Council is freely available in the ISME Journal: http://www.nature.com/ismej/journal/vaop/ncurrent/full/ismej2014213a.html doi: 10.1038/ismej.2014.213
Publication date: 9 February 2015
